Overview
Each model is associated with an initial state, parameter and simulation function. If you are using a model with the code * we pass the parameters for the ODE system to the *_param function, the initial compartment states to *_state0 and finally run the system using simulate_*. Models named *_c are age-structured, and require a contact matrix and age distribution. These are also the most mature models, all models come with a lifecycle badge making clear the potential for future changes.
Homogeneous mixing
These models assume the contact rates between individuals in the population can be adequately described by a single value, rather than varying by age or setting.
SIR model
The key functions for this model are sir_state0 sir_param and simulate_sir.
Figure 1: SIR model forward dynamics
Figure 2: SIR model infection dynamics
# A minimal example
param <- sir_param(R0 = 2.5,gamma = 0.1)
state0 <- sir_state0(S = 100,I = 1,R = 0)
res <- simulate_sir(t = 100,state_t0 = state0,param = param)
plot(res,y=c("S","I","R"),main="SIR model")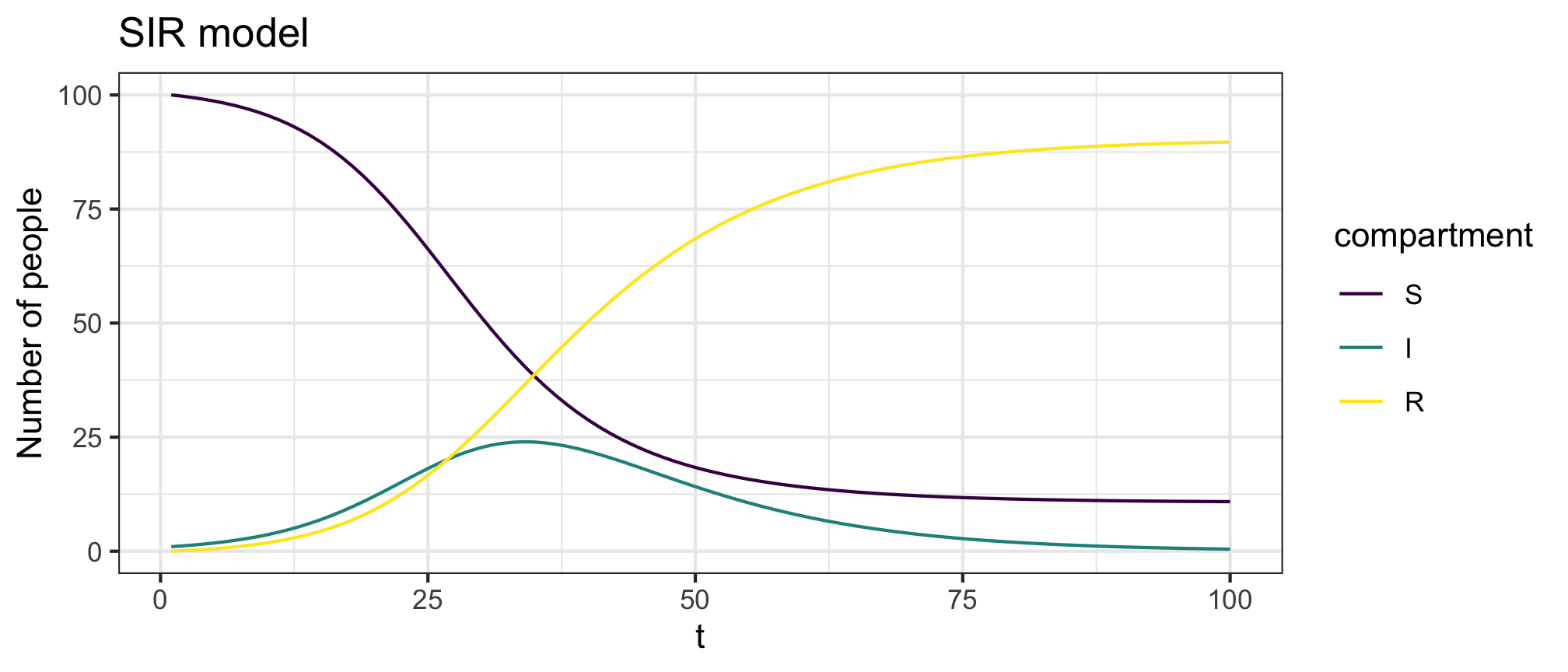
SEIR model
The key functions for this model are seir_state0 seir_param and simulate_seir.
Figure 3: SEIR model forward dynamics
Figure 4: SEIR model infection dynamics
# A minimal example
param <- seir_param(R0 = 2.5,gamma = 0.1,sigma=0.1)
state0 <- seir_state0(S = 100,E = 1, I = 0,R = 0)
res <- simulate_seir(t = 200,state_t0 = state0,param = param)
plot(res,y=c("S","E","I","R"),main="SEIR model")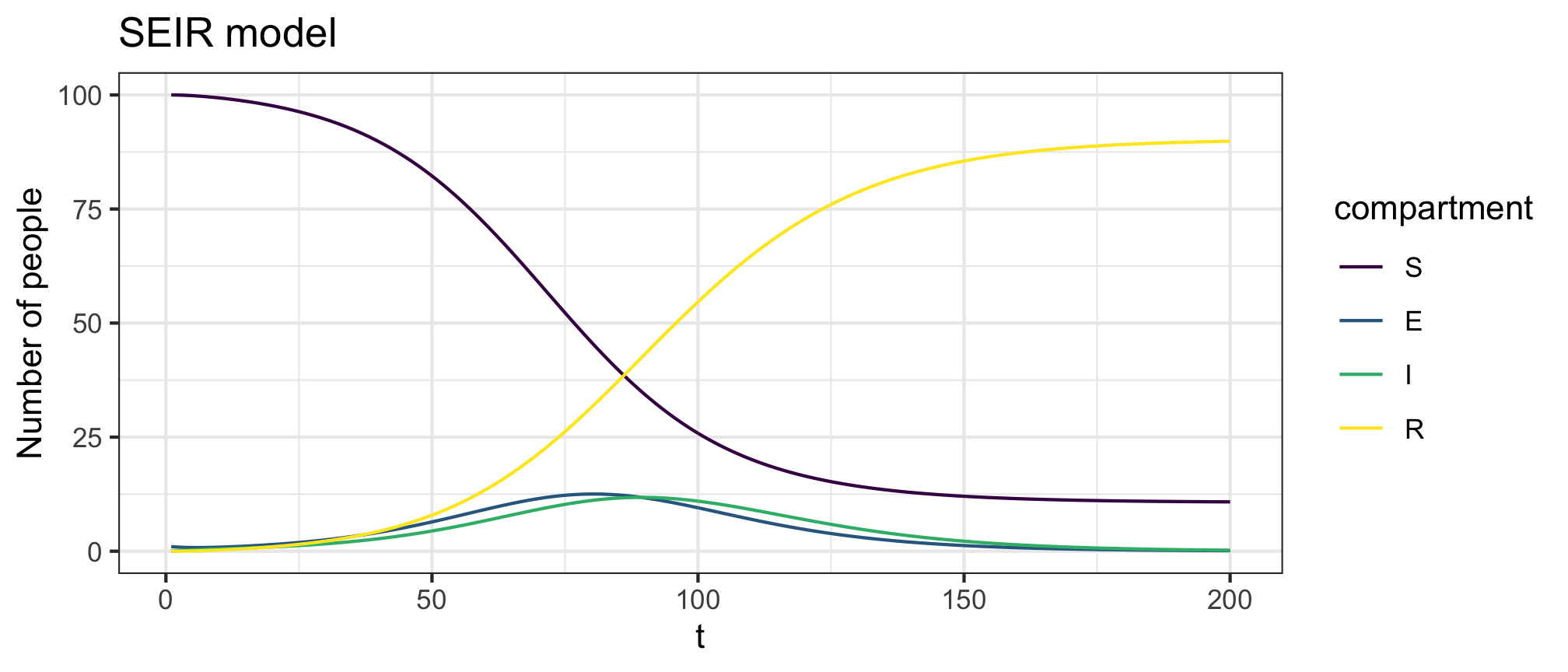
Expanded SEIR model I
This model is recommended for modelling COVID-19, it is based on 1 and 2.
The key functions for this model are seir1_state0 seir1_param and simulate_seir1.
Figure 5: Expanded SEIR model I forward dynamics
Figure 6: Expanded SEIR model I infection dynamics
# A minimal example
state0 <- seir1_state0(S = 1e5, E1 = 90, E2 = 40)
param <- seir1_param(R0=2.5,sigma1=0.2,sigma2=0.2,gamma1=0.2,gamma2=0.2,gamma3=0.2,
Qeff=0.5,Heff=0.9,rho=0.1,alpha=0.1,eta=0.02)
res <- simulate_seir1(t = 250,state_t0 = state0,param = param)
plot(res,y=c("S","E","I","Recov","Fatal"),main="Expanded SEIR model I")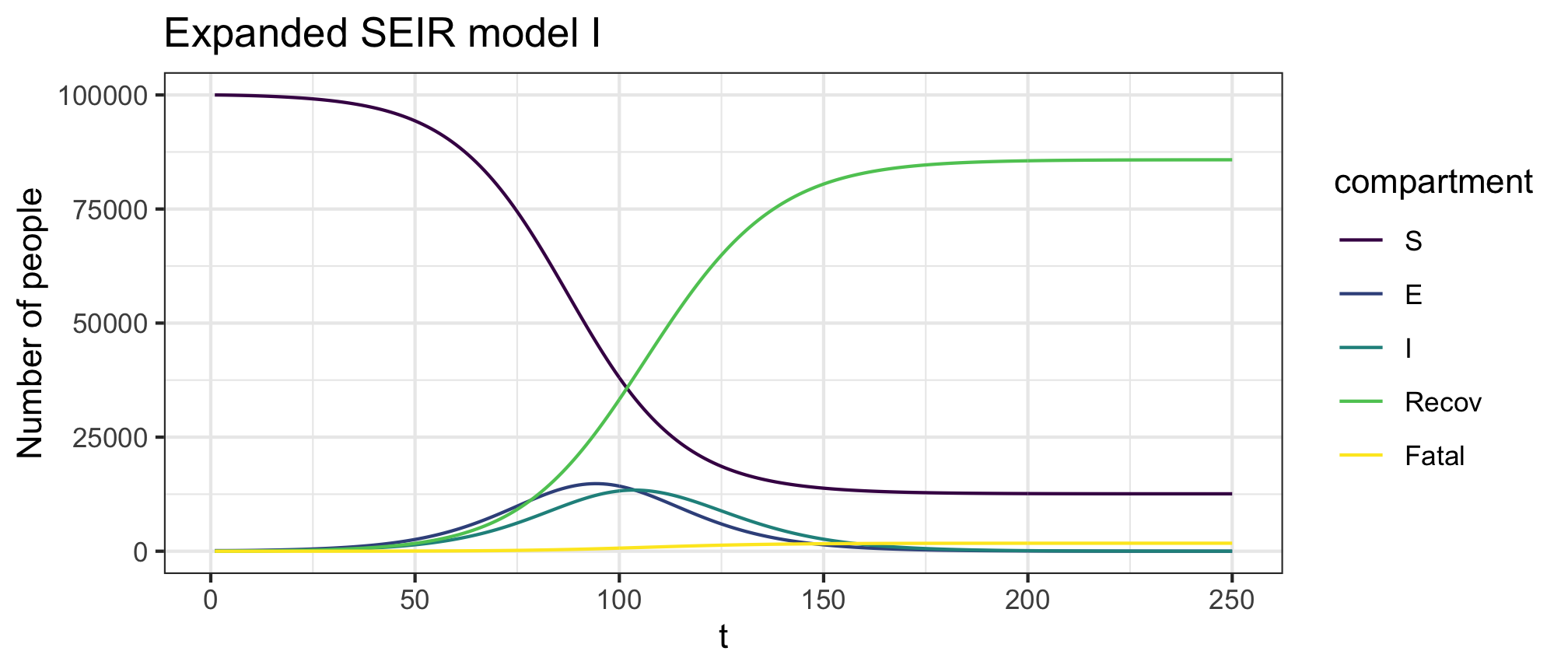
Expanded SEIR model II
This model is recommended for modelling COVID-19, it is a (partial) reproduction of 2.
The key functions for this model are seir2_state0 seir2_param and simulate_seir2.
Figure 7: Expanded SEIR model II (Doherty model) forward dynamics
Figure 8: Expanded SEIR model II (Doherty model) infection dynamics
# A minimal example
state0 <- seir2_state0(S = 1e5, E1 = 90, E2 = 40)
param <- seir2_param(R0=2.5,lambdaimp=0,sigma1=0.2,sigma2=0.2,gamma1=0.2,gamma2=0.2,
gammaq1=0.1,gammaq2=0.1,Qeff=0.5,Meff=0.99,rho=0.0,eta=1/sqrt(2),
alphamBeta=0.5,probHospGivenInf=0.09895,delta=1/14,kappa=20,pm=1)
res <- simulate_seir2(t = 250,state_t0 = state0,param = param)
plot(res,y=c("S","E","I","Recov","Fatal"),main="Moss et al (2020) model")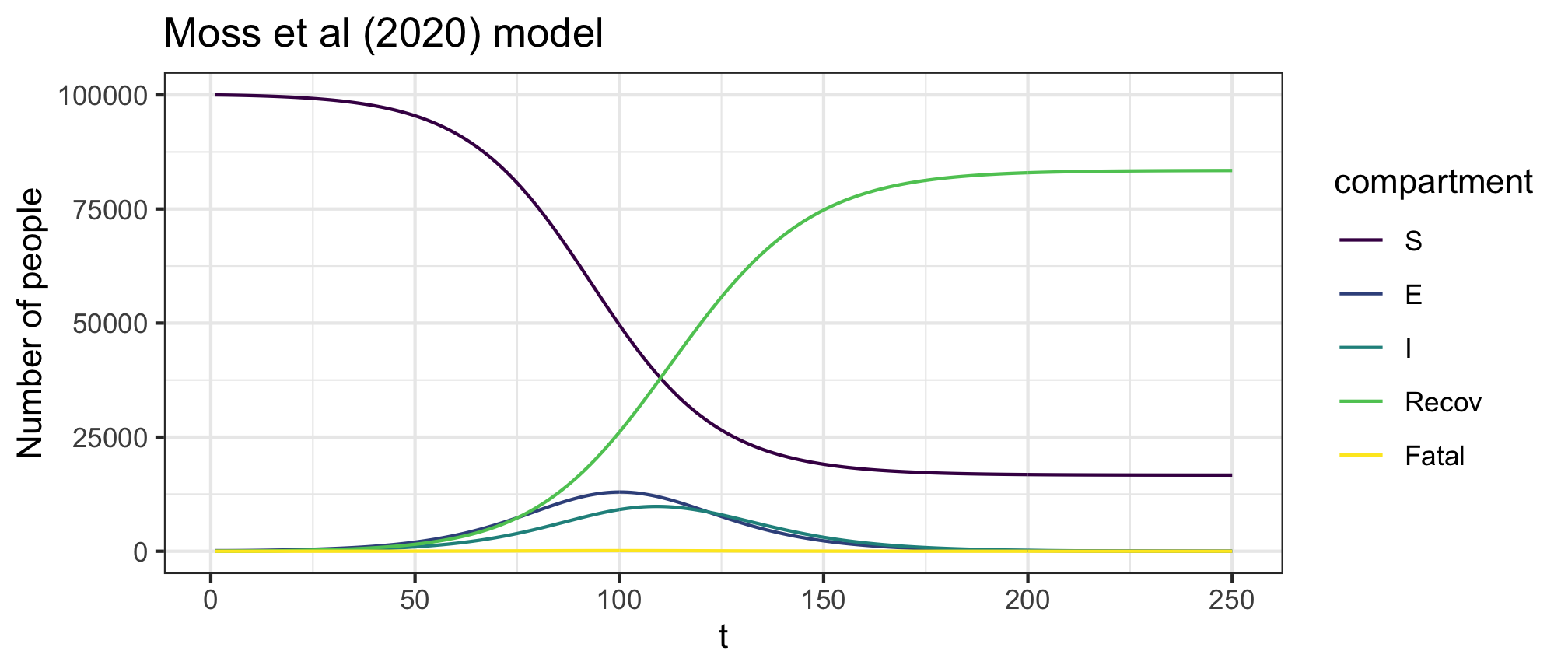
Age structured mixing
These models structure the population by discrete age partitions, with the contact rate assumed to be age and setting dependent (see figure).
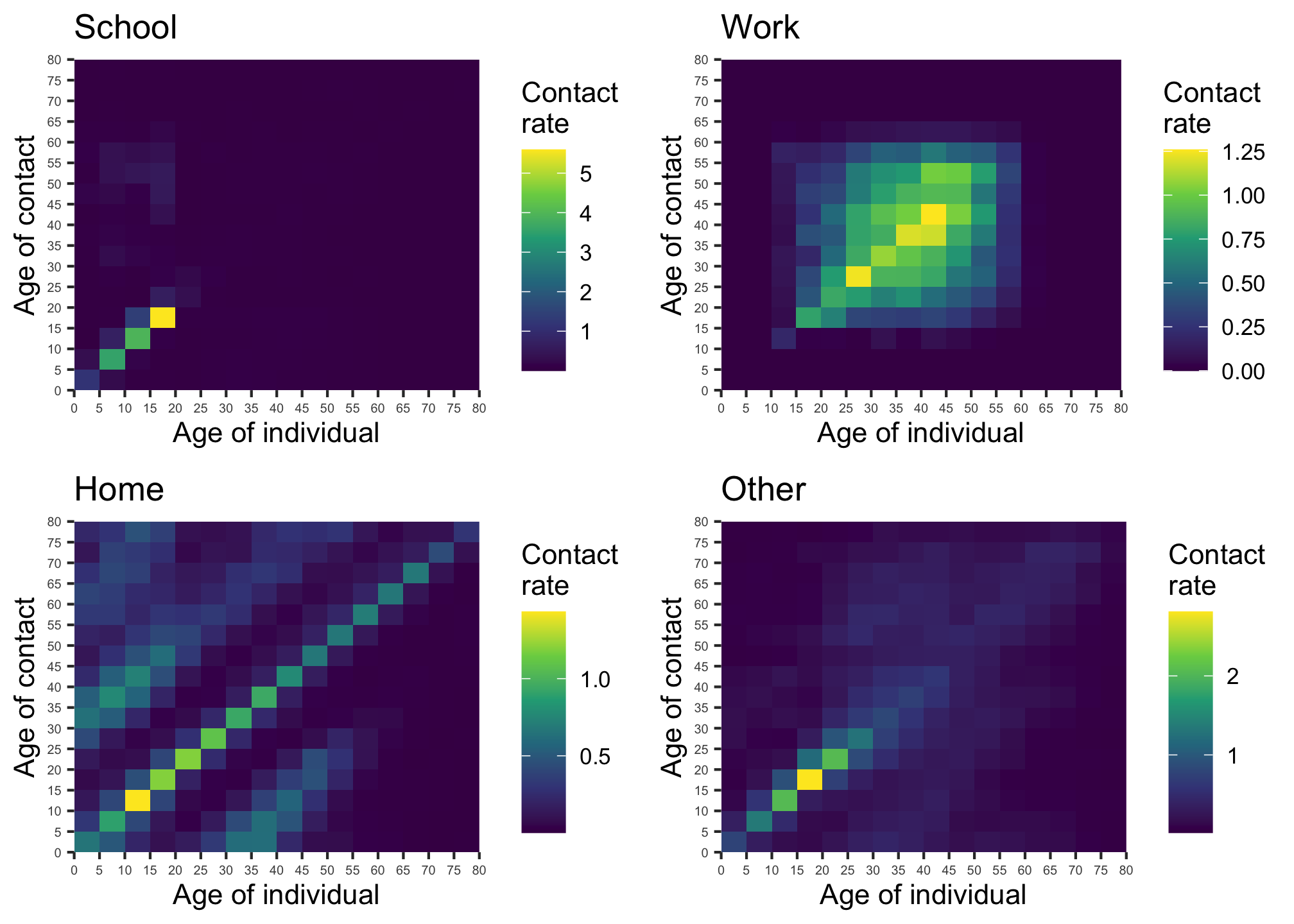
Figure 9: Contact matrices for school, work, home and other locations (source: Prem, Cook and Jit (2017)). Note the differing scales.
SIR model
The key functions for this model are sir_c_state0 sir_c_param and simulate_sir_c.
Figure 10: Age structured SIR model forward dynamics. There are j=1,…,J age groups.
Figure 11: Age structured SIR model infection dynamics. There are j=1,…,J age groups.
# A minimal example
cm_oz <- import_contact_matrix("Australia","general")
dist_oz <- import_age_distribution("Australia")
param <- sir_c_param(R0 = 2.5,gamma = 0.1,cm=cm_oz,dist=dist_oz)
nJ <- ncol(cm_oz)
S <- rep(100,nJ)
I <- rep(1,nJ)
R <- rep(0,nJ)
state0 <- sir_c_state0(S = S,I = I,R = R)
res <- simulate_sir_c(t = 150,state_t0 = state0,param = param)
plot(res,y=c("S","I","R"),main="Age structured SIR model")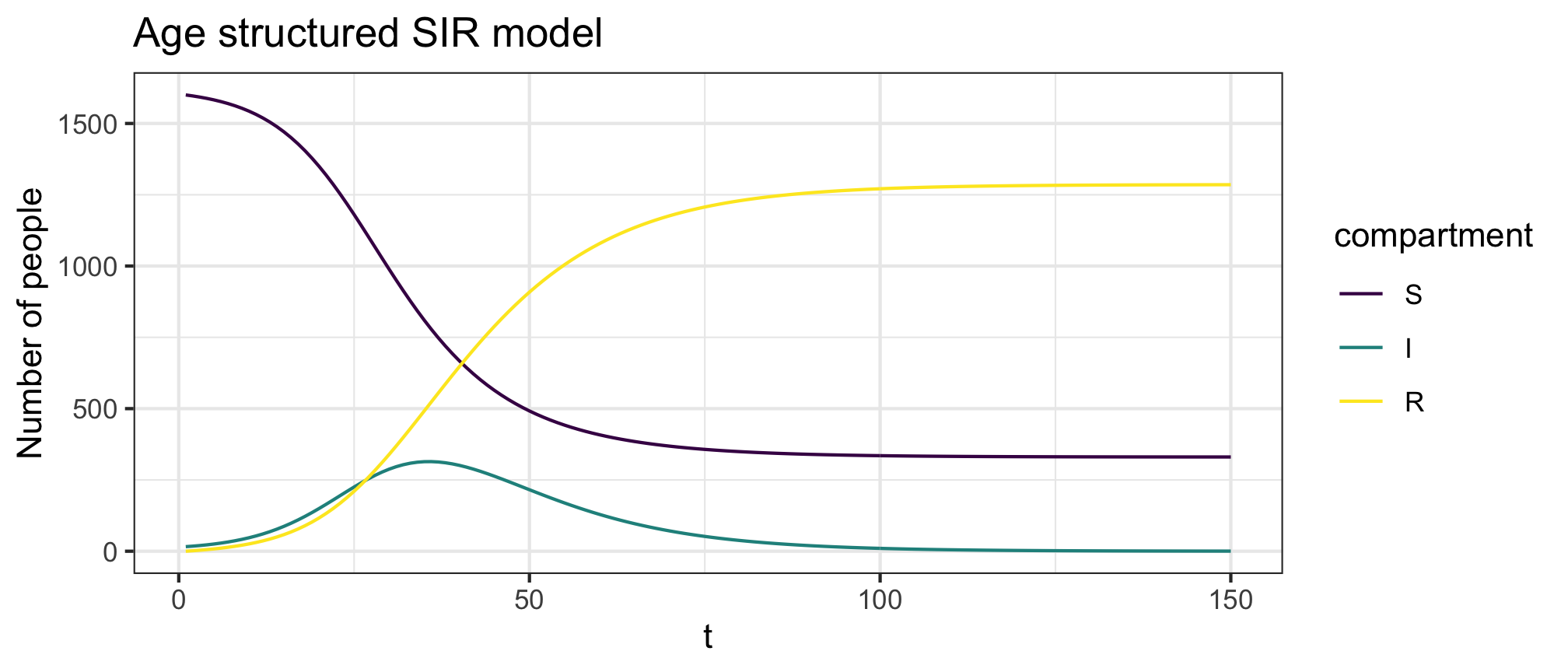
SEIR model
The key functions for this model are seir_c_state0 seir_c_param and simulate_seir_c.
Figure 12: Age structured SEIR model forward dynamics. There are j=1,…,J age groups.
Figure 13: Age structured SEIR model infection dynamics. There are j=1,…,J age groups.
# A minimal example
cm_oz <- import_contact_matrix("Australia","general")
nJ <- ncol(cm_oz)
dist_oz <- import_age_distribution("Australia")
S <- rep(1000,nJ)
E <- rep(1,nJ)
I <- rep(0,nJ)
R <- rep(0,nJ)
state0 <- seir_c_state0(S = S,E = E,I = I,R = R)
param1 <- seir_c_param(R0 = 2.5,sigma=0.1,gamma = 0.1,cm=cm_oz,dist=dist_oz)
res1 <- simulate_seir_c(t = 200,state_t0 = state0,param = param1)
plot(res1,y=c("S","E","I","R"),main="Age structured SEIR model")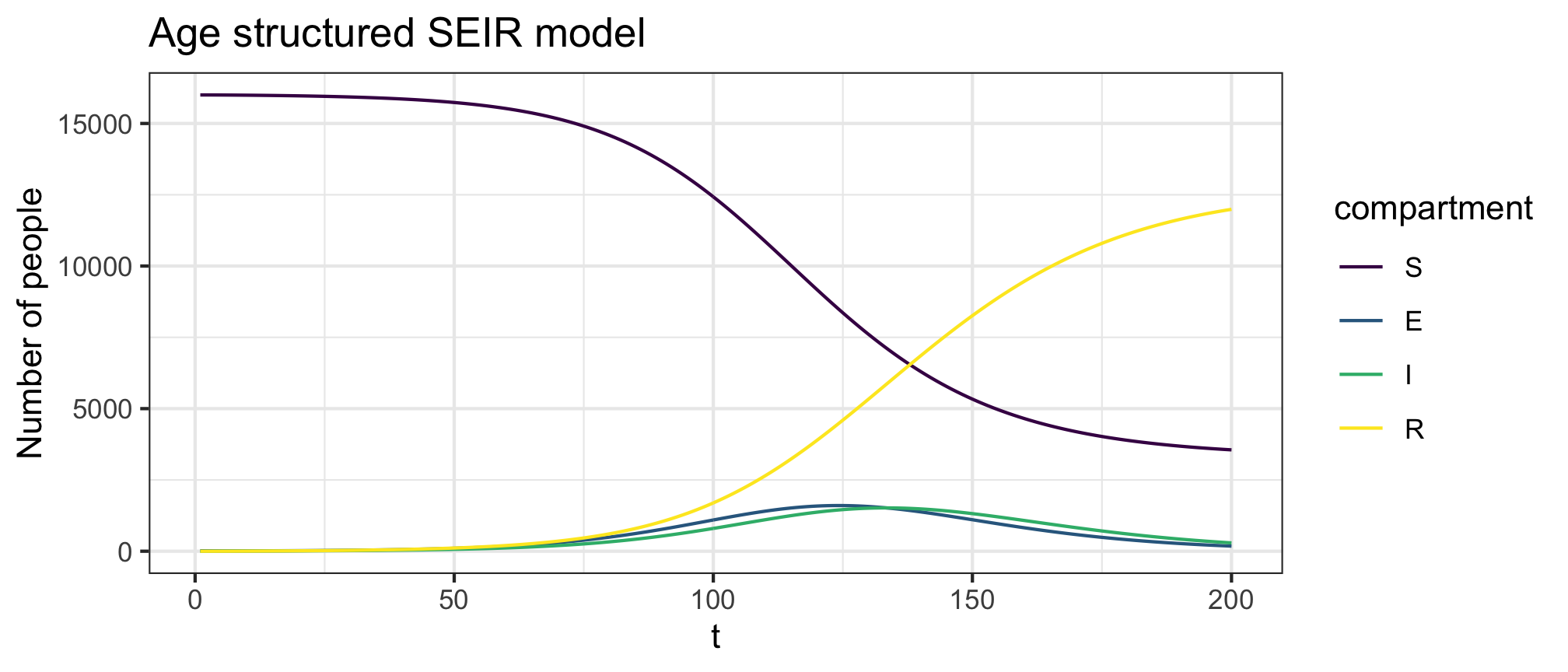
Acknowledgments
All graphs created using DiagrammR
1. Churches T, Jorm L. COVOID: A flexible, freely available stochastic individual contact model for exploring covid-19 intervention and control strategies (preprint). JMIR Preprints. Published online 2020.
2. Moss R, Wood J, Brown D, et al. Modelling the impact of covid-19 in australia to inform transmission reducing measures and health system preparedness. medRxiv. Published online 2020.